RNA Modification and Processing
Willkommen beim Transregio 319 RMaP
Was ist RMaP?
Der Transregio 319 RMaP vereint als Forschungsgemeinschaft die folgenden Schwerpunkte:
Molekularbiologie, Biochemie, Zellbiologie, chemische Biologie, biophysikalische Chemie, Strukturbiologie, Entwicklungsbiologie, Genetik und Bioinformatik.
Struktur von RMaP
A „Modifications look for Processing“
B „Processing looks for Modifications“
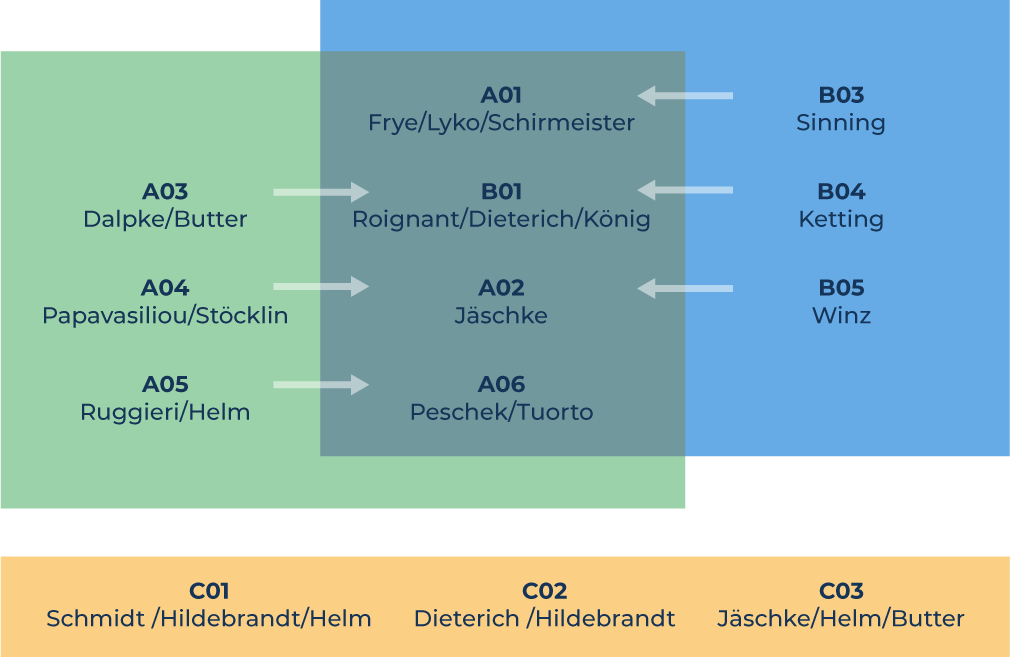
C „New RNA Technology for RMaP“
TRR Sprecherbüro

Prof. Dr. Mark Helm
Sprecher

Dr. Marion Stemke
Koordinator

Claudia Alihodzic
Administration TRR319

Doris Ebeling
Administration & Gleichberechtigung

Doris Gebhardt
Administration & Gleichstellung
Das Scientific Board von RMaP

Aktuelle Publikationen des TRR 319
Sun Y, Piechotta M, Naarmann-de Vries I, Dieterich C, Ehrenhofer-Murray AE. Detection of queuosine and queuosine precursors in tRNAs by direct RNA sequencing. Nucleic Acids Res. 2023 Oct 9:gkad826. doi: 10.1093/nar/gkad826. Epub ahead of print. PMID: 37811872.
Podvalnaya N, Bronkhorst AW, Lichtenberger R, Hellmann S, Nischwitz E, Falk T, Karaulanov E, Butter F, Falk S, Ketting RF. piRNA processing by a trimeric Schlafen-domain nuclease. Nature. 2023 Oct;622(7982):402-409. doi: 10.1038/s41586-023-06588-2. Epub 2023 Sep 27. PMID: 37758951; PMCID: PMC10567574.
Wierczeiko A, Pastore S, Mündnich S, Busch AM, Dietrich V, Helm M, Butto T, Gerber S. NanopoReaTA: a user-friendly tool for nanopore-seq real-time transcriptional analysis. Bioinformatics. 2023 Aug 7:btad492. doi: 10.1093/bioinformatics/btad492. Epub ahead of print. PMID: 37549052.
García-Vílchez R, Añazco-Guenkova AM, Dietmann S, López J, Morón-Calvente V, D’Ambrosi S, Nombela P, Zamacola K, Mendizabal I, García-Longarte S, Zabala-Letona A, Astobiza I, Fernández S, Paniagua A, Miguel-López B, Marchand V, Alonso-López D, Merkel A, García-Tuñón I, Ugalde-Olano A, Loizaga-Iriarte A, Lacasa-Viscasillas I, Unda M, Azkargorta M, Elortza F, Bárcena L, Gonzalez-Lopez M, Aransay AM, Di Domenico T, Sánchez-Martín MA, De Las Rivas J, Guil S, Motorin Y, Helm M, Pandolfi PP, Carracedo A, Blanco S. METTL1 promotes tumorigenesis through tRNA-derived fragment biogenesis in prostate cancer. Mol Cancer. 2023 Jul 29;22(1):119. doi: 10.1186/s12943-023-01809-8. PMID: 37516825; PMCID: PMC10386714.
Koch J, Neuberger M, Schmidt-Dengler M, Xu J, Carneiro VC, Ellinger J, Kriegmair MC, Nuhn P, Erben P, Michel MS, Helm M, Rodríguez-Paredes M, Nientiedt M, Lyko F. Reinvestigating the clinical relevance of the m6A writer METTL3 in urothelial carcinoma of the bladder. iScience. 2023 Jul 11;26(8):107300. doi: 10.1016/j.isci.2023.107300. PMID: 37554463; PMCID: PMC10405067.
Iyer KV, Müller M, Tittel LS, Winz ML. Molecular Highway Patrol for Ribosome Collisions. Chembiochem. 2023 Jun 29:e202300264. doi: 10.1002/cbic.202300264. Epub ahead of print. PMID: 37382189.
Yan L, Yin Z, Zhang H, Zhao Z, Wang M, Müller A, Kallenborn F, Wichmann A, Wei Y, Niu B, Schmidt B, Liu W. RabbitQCPlus 2.0: More Efficient and Versatile Quality Control for Sequencing Data. Methods. 2023 Jun 15:S1046-2023(23)00099-3. doi: 10.1016/j.ymeth.2023.06.007. PMID: 37330158.
Fradera-Sola A, Nischwitz E, Bayer ME, Luck K, Butter F. RNA-dependent interactome allows network-based assignment of RNA-binding protein function. Nucleic Acids Res. 2023 Jun 9;51(10):5162-5176. doi: 10.1093/nar/gkad245. PMID: 37070168; PMCID: PMC10250244.
Zhang H, Song H, Xu X, Chang Q, Wang M, Wei Y, Yin Z, Schmidt B, Liu W. RabbitFX: Efficient Framework for FASTA/Q File Parsing on Modern Multi-Core Platforms. IEEE/ACM Trans Comput Biol Bioinform. 2023 Jun 5;20(3):2341-2348. doi: 10.1109/TCBB.2022.3219114. Epub 2023 Jun 5. PMID: 36327193.
Eine aktuelle Auflistung der TRR-Publikationen finden Sie hier.























