RNA Modification and Processing
Welcome to Transregio 319 RMaP
RMaP News
Save the date:
RNA Modification and Processing
5th Symposium on NUCLEIC ACID MODIFICATIONS
07-09 Sept 2026 in IMB, Mainz
Transregio 319 RMaP extended for a further period
DFG – Deutsche Forschungsgemeinschaft – DFG to Fund Nine New Collaborative Research Centres
Excellent Franco-German cooperation in the field of RNA Modification research
What is RMaP?
Transregio 319 RMaP is a joint venture combining:
Molecular Biology, Biochemistry, Cellular Biology, Chemical Biology, Biophysical Chemistry, Structural Biology, Developmental Biology, Genetics, Bioinformatics.
Structure of RMaP
A „Modificationslook for Processing“
B „Processing looks for Modifications“
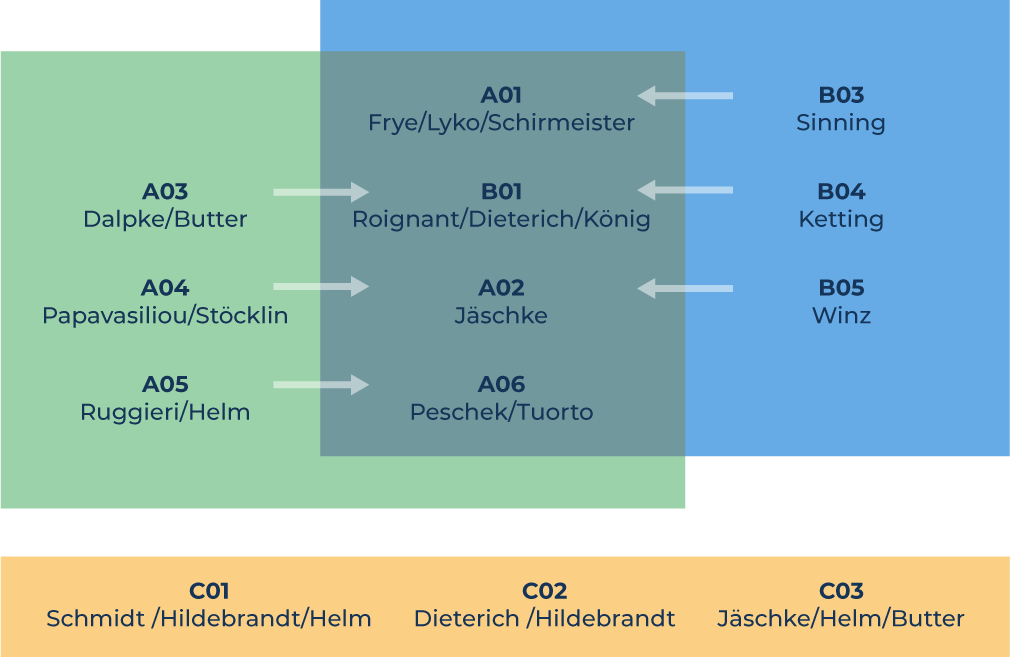
C „New RNA Technology for RMaP“
TRR Speaker Office

Prof. Dr. Mark Helm
Speaker

Dr. Marion Stemke
Executive officer and Coordinator

Claudia Alihodzic
Administrator TRR319
Members of the Scientific Board

Recent publications from the TRR 319
Wild K, Klein M, Burkard A, Marchand V, Kellner N, Paez A, Pastore S, Butto T, Motorin Y, Helm M, Sinning I. Pseudouridine increases ribosome stability in a thermophilic eukaryote. Nucleic Acids Res. 2025 Nov 26;53(22):gkaf1368. doi: 1093/nar/gkaf1368. PMID: 41432034; PMCID: PMC12723226.
Groza P, Kumari K, Esteva-Socias M, Schott J, Bhattarai DP, Sajkowska JJ, Dasgupta R, Peula C, Destefanis E, Williams C, Marchand V, Wikström P, Wiberg R, Campos AB, Gilthorpe JD, Pop B, Mateus A, Motorin Y, Dassi E, Petzold K, Tuorto F, Aguilo F. Fibrillarin-dependent 2′-O-methylation modulates RPS28 ribosome incorporation and oncogenic translation. Cancer Lett. 2025 Nov 17:218124. DOI: 1016/j.canlet.2025.218124. Online ahead of print. PMID: 41260515.
Tuorto F, Lyko F. Microbial metabolites shape mammalian protein translation. Cell Metab. 2025 Dec 2;37(12):2301-2302. doi: 1016/j.cmet.2025.11.003. PMID: 41338177.
Pauli C, Kienhöfer M, Blank MF, Begik O, Rohde C, Zimmermann SMN, Werner L, Heid D, Xu F, Weidenauer K, Delaunay S, Krall N, Trunk K, Zhao D, Zhou F, Llovera L, Ollivier A, Heit-Mondrzyk A, Platzbecker U, Baldus C, Serve H, Bornhäuser M, Vågbø CB, Benitah SA, Krijgsveld J, Novoa EM, Müller-Tidow C, Frye M. Disrupting tRNA modifications to target mitochondrial vulnerabilities in drug-resistant leukemia cells. Blood. 2025 Nov 13;146(20):2443-2456. doi: 1182/blood.2024027822. PMID: 40749163.
Tseduliak V-M, Koshla O, Mulartschyk SN, Marchand V, Motorin Y, Helm M, Ostash B. Probing the function of Streptomyces albidoflavusJ1074 gene XNR_5296 for SPOUT family ribose methyltransferase. Microbiol Spectr. 2025 Nov 26:e0219225. doi: 1128/spectrum.02192-25. Epub ahead of print. PMID: 41296997.
Zupfer K, Kaduhr L, Bessler L, Helm M, Schaffrath R. Evolutionary conservation of ubiquitin-like protein urmylation as revealed by URM1 gene shuffle from archaea to yeast. Commun Biol. 2025 Nov 23;8(1):1637. doi: 1038/s42003-025-09212-3. PMID: 41276627; PMCID: PMC12645042.
Weber M, Raorane K, Grampp CJ, Bourguignon V, Kilz LM, Glänzer D, Marchand V, Kreutz C, Motorin Y, Helm M. Mapping of HOCl-oxidized RNA identifies abasic sites as major damage and oxidation product of oxo8G. Nat Commun. 2025 Nov 21;16(1):10251. doi: 1038/s41467-025-65108-0. PMID: 41271707; PMCID: PMC12639000.
Köhler S, Peschek J. Interdomain assembly between the fungal tRNA ligase adenylyltransferase and kinase domain. RNA. 2025 Nov 17;31(12):1800-1811. doi: 10.1261/rna.080592.125. PMID: 40983467; PMCID: PMC12621585
Kallenborn F, Chacon A, Hundt C, Sirelkhatim H, Didi K, Cha S, Dallago C, Mirdita M, Schmidt B, Steinegger M. GPU-accelerated homology search with MMseqs2. Nat Methods. 2025 Sep 18. https://doi.org/10.1038/s41592-025-02819-8.. Epub ahead of print. PMID: 40968302.
Hoba SN, Schwickert M, Kammerer L, Sabin M, Weldert AC, Nidoieva Z, Meidner JL, Barthels F, Schirmeister T, Kersten C. Parallel synthesis of 5′-amino-5′-deoxy-adenosine derivatives for focused chemical space exploration and their application as methyltransferase inhibitors. RSC Med Chem. 2025 Aug 12. doi: 10.1039/d5md00376h. Epub ahead of print. PMCID: PMC12412055; DOI: 1039/d5md00376h.
Frey AF, Schwan M, Weldert AC, Kadenbach V, Kopp J, Nidoieva Z, Zimmermann RA, Gleue L, Zimmer C, Jörg M, Friedland K, Helm M, Sinning I, Barthels F. DNA-encoded library screening uncovers potent DNMT2 inhibitors targeting a cryptic allosteric binding site. iScience. 2025 Aug 5;28(9):113300. doi: 10.1016/j.isci.2025.113300. PMCID: PMC12396291; DOI: 1016/j.isci.2025.113300
van der Toorn W, Naarmann-de Vries IS, Liu-Wei W, Dieterich C, von Kleist M. WarpDemuX-tRNA: barcode multiplexing for nanopore tRNA sequencing. Nucleic Acids Res. 2025 Sep 5;53(17):gkaf873. doi: 10.1093/nar/gkaf873. PMID: 40930527; DOI: 1093/nar/gkaf873
Naarmann-De Vries IS, Preissendörfer T, König J, Dieterich C. AvaII senses m6A and inosine sites and enables targeted nanopore direct RNA-sequencing. Front Mol Biosci. 2025 Jul 28;12:1593637. DOI: 3389/fmolb.2025.1593637. PMID: 40791658; PMCID: PMC12336230.
Landrock MF, Krutyhołowa R, Böhnert P, Mazur J, Honc M, Hammermeister A, Bessler L, Scherf D, Elms A, Radczuk N, Skupien-Rabian B, Jankowska U, Herberg FW, Helm M, Klassen R, Glatt S, Schaffrath R. Autophosphorylation of conserved yeast and human casein kinase 1 isozymes regulates Elongator-dependent tRNA modifications. Nucleic Acids Res. 2025 Sep 5;53(17):gkaf881. DOI: 1093/nar/gkaf881. PMID: 40930532.
Peschek J, Tuorto F. Interplay Between tRNA Modifications and Processing. J Mol Biol. 2025 Aug 15;437(16):169198. DOI: 1016/j.jmb.2025.169198. Epub 2025 May 22. PMID: 40404521.
Kopietz K, Raorane K, Guo W, Flegler F, Bourguignon V, Thuillier Q, Kilz LM, Weber M, Marchand V, Reuter K, Tuorto F, Helm M, Motorin Y. TGT Damages its Substrate tRNAs by the Formation of Abasic Sites in the Anticodon Loop. J Mol Biol. 2025 Aug 15;437(16):169000. DOI: 10.1016/j.jmb.2025.169000. Epub 2025 Feb 26. PMID: 40011082.
Köhler S, Kopp J, Maiti S, Bujnicki JM, Peschek J. Structure of fungal tRNA ligase Trl1 with RNA reveals conserved substrate-binding principles. Nat Struct Mol Biol. 2025 Jun 25. DOI: 1038/s41594-025-01589-3. Epub ahead of print. PMID: 40563009.
Müller A, Wichmann A, Kallenborn F, Hildebrandt A, Schmidt B. RMapAlign3N: Fast mapping of 3N-Reads, Bioinformatics Advances, 2025; vbaf164. https://doi.org/10.1093/bioadv/vbaf164
Alagna N, Mündnich S, Miedema J, Pastore S, Lehmann L, Wierczeiko A, Friedrich J, Walz L, Jörg M, Butto T, Friedland K, Helm M, Gerber S. ModiDeC: a multi-RNA modification classifier for direct nanopore sequencing. Nucleic Acids Res. 2025 Jul 19;53(14):gkaf673. PMCID: PMC12276011 DOI: 1093/nar/gkaf673.
Hewel C, Wierczeiko A, Miedema J, Hofmann F, Weissbach S, Dietrich V, Friedrich J, Holthoefer L, Haug V, Mündnich S, Schartel L, Jenson K, Diederich S, Sys S, Butto T, Paul N, Koch J, Lyko F, Kraft F, Schweiger S, Lemke EA, Helm M, Linke M, Gerber S: Direct RNA sequencing (RNA004) allows for improved transcriptome assessment and near real-time tracking of methylation for medical applications https://doi.org/10.1101/2024.07.25.605188
Weldert AC, Frey AF, Krone MW, Krähe F, Kuhn H, Kersten C, Barthels F. Structure-guided design of a methyltransferase-like 3 (METTL3) proteolysis targeting chimera (PROTAC) incorporating an indole-nicotinamide chemotype. RSC Med Chem. 2025 Jun 19. doi: 10.1039/d5md00359h. Epub ahead of print. PMID: 40599585; PMCID: PMC12207533 DOI: 1039/d5md00359h
Guo W, Kaczmarczyk I, Kopietz K, Flegler F, Russo S, Cigirgan E, Chramiec-Głąbik A, Koziej Ł, Cirzi C, Peschek J, Reuter K, Helm M, Glatt S, Tuorto F. Queuosine is incorporated into precursor tRNA before splicing. Nat Commun. 2025 Jul 31;16(1):7044. doi: 10.1038/s41467-025-62220-z. PMCID: PMC12313893 DOI: 1038/s41467-025-62220-z
Ćorović M, Hoch-Kraft P, Zhou Y, Hallstein S, König J, Zarnack K. m6A in the coding sequence: linking deposition, translation, and decay. Trends Genet. 2025 Jul 7:S0168-9525(25)00132-5. DOI: 10.1016/j.tig.2025.06.002. Epub ahead of print. PMID: 40628588.
Spangenberg J, Mündnich S, Busch A, Pastore S, Wierczeiko A, Goettsch W, Dietrich V, Pryszcz LP, Cruciani S, Novoa EM, Joshi K, Perera R, Di Giorgio S, Arrubarrena P, Tellioglu I, Poon CL, Wan YK, Göke J, Hildebrandt A, Dieterich C, Helm M, Marz M, Gerber S, Alagna N. The RMaP challenge of predicting RNA modifications by nanopore sequencing. Commun Chem. 2025 Apr 12;8(1):115. doi: 10.1038/s42004-025-01507-0. PMID: 40221591;PMCID: PMC11993749.
Teschner D, Gomez-Zepeda D, Łącki MK,Kemmer T, Busch A, Tenzer S, Hildebrandt A. Rustims: An Open-Source Framework for Rapid Development and Processing of timsTOF Data-Dependent Acquisition Data. J Proteome Res. 2025 May 2;24(5):2358-2368. doi: 10.1021/acs.jproteome.4c00966. Epub 2025 Apr 22. PMID: 40260647;PMCID: PMC12053931.
Scherf D, Hammermeister A, Böhnert P, Burkard A, Helm M, Glatt S, Schaffrath R. tRNA binding to Kti12 is crucial for wobble uridine modification by Elongator. Nucleic Acids Res. 2025 Apr 10;53(7):gkaf296. doi: 10.1093/nar/gkaf296. PMID: 40226916; PMCID: PMC11995267.
Kopietz K, Raorane K, Guo W, Flegler F, Bourguignon V, Thuillier Q, Kilz LM, Weber M, Marchand V, Reuter K, Tuorto F, Helm M, Motorin Y. TGT Damages its Substrate tRNAs by the Formation of Abasic Sites in the Anticodon Loop. J Mol Biol. 2025 Feb 25:169000. DOI: 1016/j.jmb.2025.169000. Epub ahead of print. PMID: 40011082.
Gonzalez-Jabalera P, Jäschke A. Flavin adenine dinucleotide (FAD) as a non-canonical RNA cap: Mechanisms, functions, and emerging insights. Arch Biochem Biophys. 2025 Apr;766:110326. DOI: 10.1016/j.abb.2025.110326. Epub 2025 Feb 5. PMID: 39921141.
Nidoieva Z, Sabin MO, Dewald T, Weldert AC, Hoba SN, Helm M, Barthels F. A microscale thermophoresis-based enzymatic RNA methyltransferase assay enables the discovery of DNMT2 inhibitors. Commun Chem. 2025 Feb 3;8(1):32. doi: 10.1038/s42004-025-01439-9. PMCID: PMC11790956 DOI: 1038/s42004-025-01439-9
Morishima T, Fakruddin M, Kanamori Y, Masuda T, Ogawa A, Wang Y, Schoonenberg VAC, Butter F, Arima Y, Akaike T, Moroishi T, Tomizawa K, Suda T, Wei FY, Takizawa H. Mitochondrial translation regulates terminal erythroid differentiation by maintaining iron homeostasis. Sci Adv. 2025 Feb 21;11(8):eadu3011. Epub 2025 Feb 21. PMID: 39983002; PMCID: PMC11844735 DOI: 1126/sciadv.adu3011
He YM, Burkard A, Wu CS, Jin XY, Liu L, Helm M, Cheng L. A Direct Chemical Repair of Etheno-Type RNA Damages. Angew Chem Int Ed Engl. 2025 Jan 15;64(3):e202415417. DOI: 1002/anie.202415417 Epub 2024 Dec 2. PMID: 39589841.
Xu J, Koch J, Schmidt C, Nientiedt M, Neuberger M, Erben P, Michel MS, Rodríguez-Paredes M, Lyko F. Loss of YTHDC1 m6A reading function promotes invasiveness in urothelial carcinoma of the bladder. Exp Mol Med. 2025 Feb;57(1):118-130. DOI: 1038/s12276-024-01377-x. Epub 2025 Jan 1. PMID: 39741187; PMCID: PMC11799412.
Soni K, Horvath A, Dybkov O, Schwan M, Trakansuebkul S, Flemming D, Wild K, Urlaub H, Fischer T, Sinning I. Structures of aberrant spliceosome intermediates on their way to disassembly. Nat Struct Mol Biol. 2025 Jan 20. DOI: 10.1038/s41594-024-01480-7. Epub ahead of print. PMID: 39833470.
A continuously updated publication list can be found here.































