Project Area C: "New RNA Technology for RMaP"
RMaP is centered in the life sciences and embraces timely approaches and methodologies leading to fundamental insights in RNA biology. Area C is devoted to the development and maintenance of enabling techniques deemed crucial to RMaP’s effort. In comparison to other RNA related research clusters, RMaP, for obvious reasons, needs to develop and maintain particular competences with respect to RNA modification analytics. Three focus areas are embodied in the projects C01–C03, which concentrate on sequencing, modification analytics, interaction mapping, and data management. Application of the cross-cutting C technologies provides central competences to areas A and B. Further technologies and competences, such as structural biology, microscopy, synthetic organic chemistry and bioconjugate chemistry, genetics, cell biology and various model organisms, reside in the individual projects and are accessible to RMaP members on an individual basis. Particular competences that are worth mentioning include highly reputed experience in protein-RNA crosslinking techniques such as CLIP (König B01, 201), CRAC (Winz, B06), and structural biology including access to cryo-EM facilities (Sinning group).
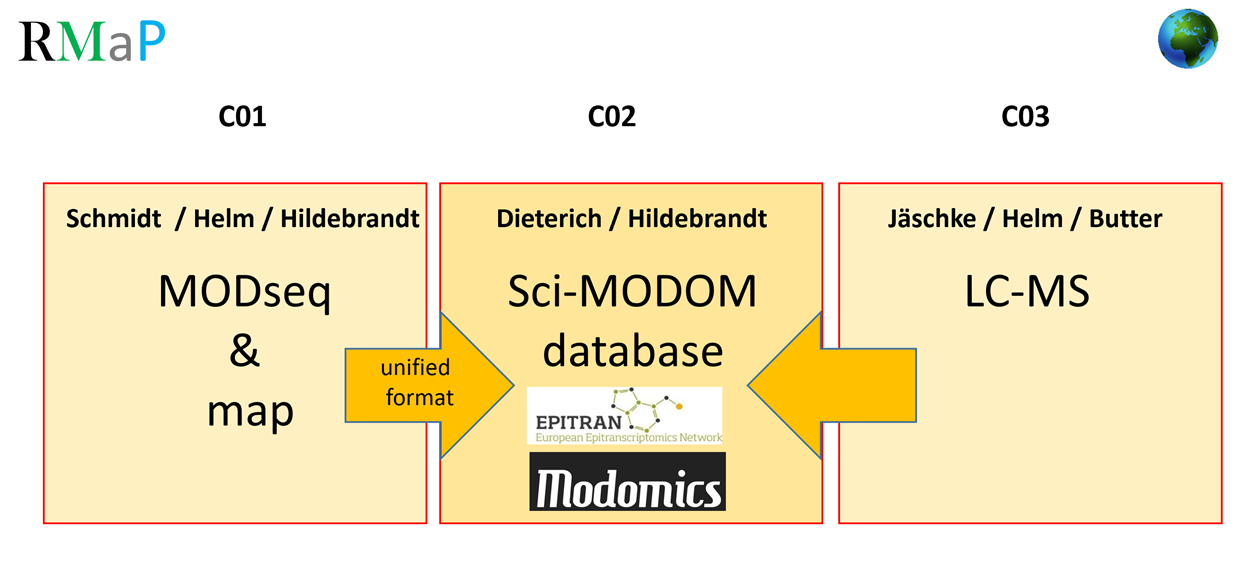
C01
Epitranscriptomics rely heavily on modification mapping techniques, requiring fast and effective treatment of massive amounts of sequencing data. New modification mapping chemistries lead to new data formats and, like conventional ones, require considerable efforts for data treatment and analysis. Here, a new chemistry will be developed along with mapping, and base calling techniques in a harmonized approach. Standards and tools for quality and performance assessment will be developed to improve accuracy and speed, including machine learning, ultra-fast implementations, and easy-to-use pipelines, which will be applicable to other mapping chemistries, as well.
C02
Availability of, and access to high throughput RNA modification data is indispensable for a global understanding of epitranscriptomics, and a mission critical factor of RMaP. Here, SCI-MODOM will be developed as the central data repository and database hub for RMaP. SCI-MODOM will provide analytics combining modification sites and processing events. A database infrastructure and computational resources will be developed, and data exchange formats, interoperability standards and interfaces developed, to facilitate data curation, integration, access and availability. This project will promote SCI-MODOM as the database solution for high-throughput assays.
C03
Mass spectrometry has become a powerful tool to study protein-RNA interactions. However, as standard analyses remain typically limited to post-translational protein modifications, very few of these approaches have been extended to modified RNA, experimentally or bioinformatically. Here, LC-MS technologies that take RNA modifications into account when characterizing RNA-protein interactions will be developed. In addition to extensive experimental Analyses of RNA and RNA-protein complexes, this also includes the screening of datasets from public repositories to systematically include RNA modifications.
